BAM Chapter 26B: Detection of Hepatitis A Virus in Foods
Bacteriological Analytical Manual (BAM) Main Page
Authors: Jacquelina Williams-Woods, Gary Hartman, and William Burkhardt III
Contact: Jacquelina Woods
Revision History:
- New BAM Chapter; October 2013; posted: January 2014.
- Corrections made to Primer and Probe sequences in Table 1 (April 2014) and Table 4 (June 2014) .
Introduction
Hepatitis A is a non-enveloped RNA virus 27 to 32 nm in diameter. It has an icosahedral symmetry and belongs to the genus Hepatovirus of the Picornaviridae family. Infections with HAV can produce effects that range in severity from asymptomatic to death from fulminant hepatitis. Infections with HAV are typically self-limiting and do not result in chronic liver disease, however, outbreaks of HAV require considerable amounts of public health resources due to long incubation and shedding period after infection The virus in shed in the feces and peak fecal excretion, hence infectivity, occurs prior to the onset of symptoms (Lednar et al., 1985). These viruses are considered to be highly infectious and illness can be caused by as few as 10 viral particles (Teunis, 2008). Illnesses caused by transmission of these viruses in food have been epidemiologically-linked to three distinct classes: (i) cases associated with the consumption of ready-to-eat (RTE) foods contaminated by food workers; (ii) cases associated with the environmental contamination of produce, and (iii) cases associated with the consumption of molluscan shellfish harvested from contaminated water.
Significant concern of food-borne viruses on food safety and public health is the current limitations on matrix-associated virus detection. Detection in implicated foods is considered difficult because of the low level of viral contamination, inefficient extraction from food matrices, and the inability to efficiently enrich viruses-an aspect beneficial to most other bacterial detection methodologies. These factors necessitate that isolated virus particles be sufficiently concentrated in order to detect their presence in foods. Such limitations have hindered the FDA’s efforts to provide effective regulatory oversight in surveillance and outbreak investigations. Advances in molecular detection techniques e.g. reverse transcription (RT)-PCR and real-time RT-qPCR (Arnal, Ferre-Aubineau et al. 1999; Schwab, Neill et al. 2000; Sair, D'Souza et al. 2002) have been shown to offer specificity and sensitivity for food-borne pathogen detection. These methods are ideal for the detection of viruses with low titers. The use of these methods during outbreaks and routine surveillance would allow for faster sample throughput by quickly screening samples and allowing for more time to analyze "Cannot Rule Out" (CRO’s). In addition, at the time of this study, there were no multi-lab validated rapid protocols available for detection of enteric viruses in food.
Detection Method
The detection assay includes oligonucleotide primers and dual-labeled hydrolysis (Taqman-style) probes for the in vitro quantifiable detection of HAV. The HAV primers are from the 5’ untranslated region (UTR) of the genome and designed for detection of all genotypes of HAV (Gardner et al. 2003). This assay also incorporates an RNA internal amplification control (IAC) to monitor any potential matrix-derived inhibitory effects.
Equipment/Supplies
- DNase/RNase-free microcentrifuge tubes, 0.5 and 1.5 mL, non-stick, low retention, siliconized
- Microcentrifuge, bench top, capable of ≥10,000 × g (capable of 0.5-2.0 mL volume)
- Ice Bucket and ice, or bench top cooler
- RNase Away® or equivalent
- Smart Cycler tube holder
- Smart Cycler cold block for Smart cycler tubes (-20 °C)
- Adjustable Micropipettors (0.2 – 1000 µl), dedicated for RNA work only
- Aerosol resistant Micropipettor tips (0.2 – 1000 µl)
- Vortex Mixer
- Smart Cycler Reaction Tubes, 25-µl capacity.
- Latex or nitrile gloves – Powder Free
- Cepheid Smart Cycler II
- Smart Cycler tube mini-centrifuge with Smart Cycler adapter
Reagents
- Qiagen OneStep RT-PCR Kit; Cat No. 210210 (25 reactions) or 210212 (100 reactions)
- OneStep Kit Components
- OneStep RT-PCR Enzyme Mix
- OneStep RT-PCR Buffer (5X)
- dNTP Mix (10mM each nucleotide
- RNase-free water
- Q-Solution (DISCARD THIS KIT COMPONENT)
- OneStep Kit Components
- 50 mM MgCl2
- Ambion Superase·In (20 units/µl); Cat No. AM2694 (2,500 U) or AM2696 (10,000 U) Life Technologies
- 10 µM Working Solutions for each primer and probe from Table 1.
- Internal Control RNA (available from BioGX Cat No. 750-0001)
- positive controls (HAV RNA—ATTC VR-1402 and murine norovirus RNA – ATCC PTA-5935)
- negative RT-qPCR control (Nuclease free-water, Applied Biosystens AM9937)
HAV Primers and Probe
All HAV probes and primers were commercially synthesized (Integrated DNA Technologies, Coralville, IA). The HAV probe is labeled 5’ with Cy5 reporter dye and 3’ with Iowa Black RQ as a quencher. The IAC probe was labeled 5’ with TxRed reporter dye and 3’ with Iowa Black RQ as a quencher. All primers and probes are hydrated in sterile primer TE buffer (see Appendix D) to 100µM concentration. Ten µM working stocks are prepared from the 100 µM stock solution and are stored at -20 °C in a frost free freezer.
Internal Control Primer, Probes and RNA
Table 1. Primer and Probe Sequences for HAV and Internal Amplification Control RNA
| Identification | Primers | Location∞ |
|---|---|---|
| GAR2F | 5’ ATA GGG TAA CAG CGG CGG ATA T 3’ | 448-469 |
| GAR1R | 5’-CTC AAT GCA TCC ACT GGA TGA G-3’ | 517-537 |
| IC46Fa,b | 5’GAC ATC GAT ATG GGT GCC G-3’ | N/A |
| IC194Ra,b | 5’-AAT ATT CGC GAG ACG ATG CAG-3’ | N/A |
| Probes | ||
| GARP | Cy5-5’ AGA CAA AAA CCA TTC AAC GCC GGA GG 3’ -IB-RQ* | 483-508 |
| IACPa,b | TxR -TCT CAT GCG TCT CCC TGG TGA ATG TG -IB RQ* | N/A
|
aInternal Amplification Control (IAC) primers and probes are covered by U.S. Patent Application 0060166232
bDepaola, Jones, Woods, et al. 2010.
∞Based on GenBank accession # M14707
*IB RQ- Iowa Black RQ
Table 2. Amplification Reaction Components
| Reagent | Initial Concentration | Volume per 25 μl reaction |
Final Concentration |
|---|---|---|---|
| RNase Free H2O | 11.05 μl | - | |
| 5 × OneStep RT-PCR Buffer | 5 × | 5.0 μl | 1 × |
| MgCl2~ | 50mM | 0.75 μl | 1.5 mM |
| dNTP Mix | 10 mM | 1 μl | 0.4 mM |
| GAR2F | 10 μM | 0.75 μl | 0.3 μM |
| GAR1R | 10 μM | 0.75 μl | 0.3 μM |
| IC 46F | 10 μM | 0.1875 μl | 0.075 μM |
| IC 194R | 10 μM | 0.1875 μl | 0.075 μM |
| GARP | 10 μM | 0.5 μl | 0.2 μM |
| IACP | 10 μM | 0.375 μl | 0.15μM |
| OneStep RT-PCR Enzyme Mix | 1.00 μl | ||
| Superase in | 20 Units/μl | 0.25 μl | 5 Units |
| Internal Amplification Control RNA | *0.2 μl | ||
| RNA | 3 µl |
* Amount varies with concentration of IAC RNA. The amount of IAC template needs to be adjusted based on the prepared stock concentration to report Cycle threshold at about 20-25 PCR cycles when no inhibition is present in the reaction. The required concentration was provided to each participating laboratory.
˜ With the addition of 1.5 mM MgCl2, the final concentration per reaction is 4.0mM MgCl2
Table 3. HAV RT-qPCR Cycling Conditions: Cepheid Smart Cycler II
| Stage 1 | Stage 2 | Stage 3 | ||||||
|---|---|---|---|---|---|---|---|---|
| Hold | Hold | 3 Temperature Cycle | ||||||
| Temp | Sec | Optics | Temp | Sec | Optics | Repeat 45 times | ||
| 50 | 3000 | Off | 95 | 900 | Off | Temp | Sec | Optics |
| 95 | 10 | Off | ||||||
| 53 | 25 | Off | ||||||
| 64 | 70 | On | ||||||
Qualitative data analysis.
On the SmartCycler II Instrument, set the following Analysis Settings for TxRd and Cy5 channels. Update analysis settings if they are changed before recording results.
- Usage: Assay
- Curve Analysis: Primary
- Threshold Setting: Manual
- Manual Threshold Fluorescence Units: 10.0
- Auto Min Cycle: 5
- Auto Max Cycle: 10
- Valid Min Cycle: 3
- Valid Max. Cycle: 60
- Background subtraction: ON
- Boxcar Avg. Cycles: 0
- Background Min. Cycle: 5
- Background Max. Cycle: 40
- Max Cycles: 50
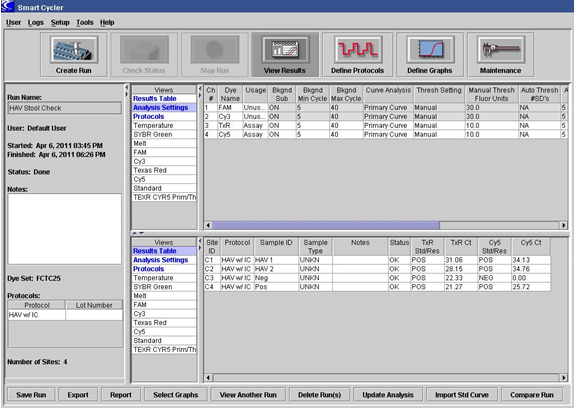
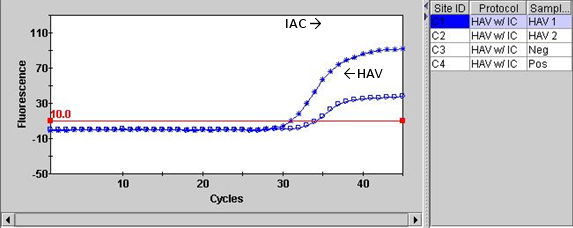
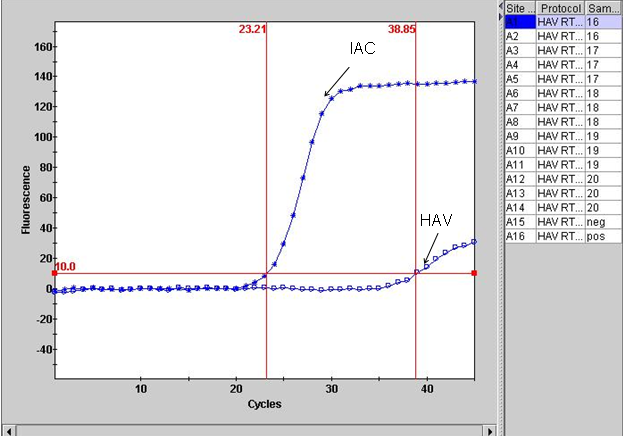
Primary fluorescence curves that cross the threshold will be recorded as "POS" and the cycle number when it crossed the threshold will be displayed in the Results Table view (Figure 1A). Negative results are shown as "NEG". The TxRd and Cy5 channels correlate to IAC and the HAV targets, respectively. Results can also be viewed graphically. For example, Figure 1B is a graphical view for the two channels for the Strain of HAV HM175/18f.
Figure 1. Example of result output from Smart Cycler II. A. Results viewed in table form, B. Graphical representation of results.
A. Analysis and Results table view
B. Results graphical view
- Negative samples:
Sample is "negative" if the RT-qPCR negative control is negative for HAV, the RT-qPCR positive control is positive for HAV, the un-spiked sample is negative for HAV, and the internal amplification control (IAC) is positive. - No further analysis is needed.
- Positive Samples:
Sample is "positive" if the RT-qPCR negative control is negative for HAV, RT-qPCR positive control is positive for HAV, the spiked HAV RNA RT-PCR sample is positive for HAV, and the internal amplification control (IAC) is positive.Note: If the negative RT-qPCR control sample demonstrates positive results crossing the Cy5 threshold or if the IAC is negative, the assay must be repeated.
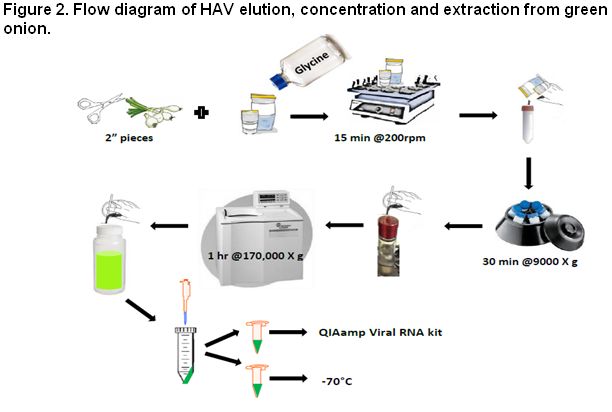
Concentration and Extraction Methods
The concentration and extraction protocol uses ultracentrifugation to concentrate HAV eluted from green onion (Figure 2) . Concentration and extraction of viruses from produce can be difficult due to the increased presence of polysaccharides. The use of the QIAshredder and a slight modification of the QIAamp viral RNA extraction kit produce a RNA viral concentrate with minimal inhibition. Murine norovirus (MNV), ATCC PTA-5935, is used as an extraction control to access the overall performance of the method.
Equipment/Supplies
- Biological Safety Cabinets (BSC- 2 )
- Latex or nitrile gloves (powder-free)
- Vortex Mixer
- Mini-centrifuge, capable of ≥2,000 × g, holds 0.5-2.0 ml microcentrifuge tubes (Labsource C90-048)
- Whirl pak bags 24 oz stand-up (Nasco B01401WA)
- Micro-centrifuge, bench top, capable of ≥10,000 × g , holds 0.5-2.0 ml microcentrifuge tubes
- Dedicated Virus, calibrated pipettors (P10, P20, P200 or equivalent)
- Dedicated RNA, calibrated pipettors (P10, P20, P200 or equivalent)
- Qiagen QIAamp Viral RNA Mini Kit Cat No. 52904 (50) or 52906 (250)
- Qiagen QIAshredder Cat No.79654 (50) or 79656 (250)
- Qiagen Collection Tubes Cat No. 19201
- Filter-Barrier aerosol resistant pipette tips
- Bench top cooling block, pre-chilled to 4°C (preferable) or Ice Bucket with Ice
- Micro-centrifuge tubes, 0.5-2.0 ml, (siliconized or low retention, certified DNase & RNase free,)
- High speed centrifuge with rotor capable of holding Falcon 50 ml tubes (or equivalent), 9,000-12,000 × g
- 50ml conical tubes or equivalent (capable of withstanding speeds ≥12,000 × g) (Fisher Scientific Cat No. 14-959-49A)
- Sorval WX Ultra Centrifuge or equivalent
- Carbon Fiber Rotor F40L, 8 × 100 ml: Cat.# 096-087057 or equivalent
- 70 mL Polycarbonate ultracentrifuge tubes w/aluminum cap tubes for F40L rotor (Cat No.010-1333 or equivalent)
- Shaker (orbital or side-to-side)
- Balance (0.01 g sensitivity)
- Sterile scissors
- Murine Norovirus (ATCC PTA-5935)
- positive controls (HAV RNA—ATCC VR-1402 and murine norovirus RNA—ATCC PTA-5935)
- negative RT-qPCR control (Nuclease free-water Applied Biosystems AM9937)
Reagents
- Glycine (Sigma G-7126 or equivalent; TLC grade or better
- Nuclease free-water Applied Biosystems (AM9937) 10 × 50ml
- NaCl(Sigma S3014 or equivalent)
- Ethanol (95-100% molecular grade) (Sigma E7023)
Concentration and Extraction work instruction details
- Add 50 g ± 2 g of produce cut in 2-5" pieces to a whirl-pak plastic bag
- Note: Include all parts of the produce present including roots to leaves.
- Add 55 mL ± 2 ml of 0.75 M Glycine Buffer (0.75 M Glycine, 0.15 M NaCl, pH 7.6 – see Appendix D), add 100 µl of extraction control (see Appendix G) and seal.
- Shake at 200rpm or medium speed for 15 min at room temperature
- Decant into a 50 mL tube (Falcon 2089 Disposable or similar) let bag sit for 2-3 min, shake side to side and pour remaining liquid into centrifuge tube
- Note: Do not squeeze bag to obtain more buffer. This action will lead to assay inhibition.
- Centrifuge at 9,000 × g for 30 min at 4 °C (2 -22 °C)
- Decant supernatant into a 50-70 mL Ultra-centrifuge tube
- Balance tubes to within 0.05g of each other using Glycine Buffer (0.75 M Glycine, 0.15 M NaCl; pH 7.6)
- minimum volume for ultracentrifugation using Fiberlite rotor and tubes is 50ml, add glycine/NaCl buffer to bring volume to ≥50ml
- Centrifuge 37,000 rpm (170,000 × g) for 60 min at 4°C (rpm speed is specific for F40L rotor, if using different rotor check speed for 170,000 × g)
- Label QIAshredder and QIAamp Spin column with sample number
- Pour (discard) off supernatant (should see pellet on side of tube), let tubes sit for 4-5 min and remove and discard remaining liquid with a micro pipettor
- Add 280 µl of glycine buffer to tube, carefully resuspend sample, and evenly distribute sample into two 1.5ml DNase/RNase free tubes. If sample pellet is larger than 0.5g, refer to Qiagen QIAamp Viral RNA Mini Kit manual instructions on how to precede with larger samples volumes. Use only 1 tube for RNA extraction. Store other tube at -70° C.
- Before starting RNA extraction place AVE elution buffer in a 70 °C heating block.
- Add 560 µl prepared (see Appendix D) Buffer AVL with carrier RNA tube
- Incubate at room temperature (15-25 °C) for 10 min
- Resuspend pellet by pipetting up and down and vortexing
- Transfer all liquid to the QIAshredder
- Centrifuge 2 min at maximum speed
- Label microcentrifuge tubes with sample number
- Carefully transfer the supernatant of the flow-through fraction to a new microcentrifuge tube without disturbing the cell-debris pellet (if present) in the collection tube
- Add 560 µL of ethanol (95-100%) to the cleared lysate, and mix immediately by pipetting. Do not centrifuge. Continue without delay to next step
- Apply 630 µl of the solution to an QIAamp mini column
- Centrifuge 8000 × g (≥10,000 rpm) for 1 min. Place QIAamp spin column in new collection tube. Discard flow through and collection tube
- Continue to add sample until the entire sample (from both microfuge tubes) has been passed through the column, discarding collection tube each time
- Add 500 µl Buffer AW1. Incubate for 10 min. Centrifuge 60 sec at 8,000 × g (10,000 rpm). Discard flow through and collection tube
- Transfer the QIAamp mini column into a new 2 ml collection tube. Pipet 500 µl Buffer AW2 onto the QIAamp Mini column. Centrifuge at full speed (20,000 × g) (14,000 rpm) for 3 min. Discard flow through and collection tube
- Transfer the QIAamp mini column into a new 2 ml collection tube. Centrifuge at full speed (20000 × g) for 1 min (14,000 rpm) to dry column
- To elute RNA, transfer the QIAamp mini column in to a new 1.5ml DNase/RNase free centrifuge tube. Pipet 35 µl of heated Buffer AVE directly onto the QiaAmp silca-gel membrane. Close the tube gently, and centrifuge for 1 min at ≥8000 × g (≥10,000 rpm)
- Add 25 µl of heated Buffer AVE to column. Pipette the eluted 35 µl back to the top of the column. Close the tube gently, and centrifuge for 1 min at ≥8000 × g (≥10,000 rpm)
- Label samples, use immediately, place elute on ice, or freeze at -70 °C for long term storage
RT-qPCR for Murine Norovirus (MNV) extraction control
This method will assess the recovery of murine norovirus from spiked samples and determine if the extraction was performed correctly. The test utilizes IAC primers and probe that are multiplexed (simultaneously amplified) with MNV primers and probe for each RNA sample. The IAC adds value to the assay by confirming that amplifiable RNA is present in samples that test PCR-negative for MNV. The IAC also verifies efficient nucleic acid extraction and removal of RT-PCR inhibitors.
Equipment/Supplies
- DNase/RNase-free microcentrifuge tubes, 0.5 and 1.5 mL, non-stick, low retention, siliconized
- Microcentrifuge (0.5-2.0 mL volume)
- Ice Bucket and ice, or bench top cooler
- RNase Away
- Smart Cycler tube holder
- Smart Cycler cold block for Smart cycler tubes (-20 °C)
- Adjustable Micropipettors (0.2 – 1000 µl), for RNA work only
- Aerosol resistant Micropipettor tips (0.2 – 1000 µl)
- Vortex Mixer
- Smart Cycler Reaction Tubes, 25-µl capacity
- RNase-free latex or nitrile gloves- Powder Free
- Cepheid Smart Cycler II
- Smart Cycler tube mini-centrifuge with Smart Cycler adapter
- MNV positive control (ATCC PTA-5935)
- Negative RT-qPCR control (Nuclease free-water Applied Biosystems AM9937)
Reagents
- Qiagen OneStep RT-PCR Kit; Cat No. 210210 (25 reactions) or 210212
(100 reactions)- OneStep Kit Components
- OneStep RT-PCR Enzyme Mix
- OneStep RT-PCR Buffer (5X)
- dNTP Mix (10mM each nucleotide)
- RNase-free water
- Q-Solution (DISCARD THIS KIT COMPONENT)
- OneStep Kit Components
- 50 mM MgCl2
- Ambion Superase In (5 units/µl); Cat No. 2694 (2,500 U) or 2696 (10,000 U)
- 10 µM Working Solutions for each primer and probe from Table 4.
- Internal Control RNA
Table 4. Primer and Probe Sequences for MNV and Internal Control RNA
| Identification | Primers | Location# |
|---|---|---|
| MNVF | 5’- TGC AAG CTC TAC AAC GAA GG -3’ | 6520-6539 |
| MNVR | 5’- CAC AGA GGC CAA TTG GTA AA 3’ | 6645-6626 |
| IC46Fa | 5’- GAC ATC GAT ATG GGT GCC G-3’ | N/A |
| IC194Ra | 5’- AAT ATT CGC GAG ACG ATG CAG-3’ | N/A |
| MNVP | Cy5- 5’ CCT TCC CGA CCG ATG GCA TC 3’-IB-RQ* | 6578-6594 |
| IACP | TxR – 5’ TCT CAT GCG TCT CCC TGG TGA ATG TG -IB RQ 3’ * | N/A |
aInternal Amplification Control (IAC) primers and probes are covered by U.S. Patent Application 0060166232
*IB RQ- Iowa Black RQ
#based on accession no. JF320650
Table 5. Amplification Reaction Components
| Reagent | Initial Concentration | Volume per 25 μl reaction |
Final Concentration |
|---|---|---|---|
| RNase Free H2O | 11.8 μl | - | |
| 5 × OneStep RT-PCR Buffer | 5 × | 5.0 μl | 1 × |
| MgCl2+ | 50 mM | 0.75 μl | 1.5 mM |
| dNTP Mix | 10 mM | 1 μl | 0.4 mM |
| MNVF | 10 μM | 0.50 μl | 0.2 μM |
| MNVR | 10 μM | 0.50 μl | 0.2 μM |
| IC 46F | 10 μM | 0.1875 μl | 0.075 μM |
| IC 194R | 10 μM | 0.1875 μl | 0.075 μM |
| MNVP | 10 μM | 0.25 μl | 0.1 μM |
| IACP | 10 μM | 0.375 μl | 0.15 μM |
| OneStep RT-PCR Enzyme Mix | 1.00 μl | ||
| Superase in | 20 Units/μl | 0.25 μl | 5 Units |
| Internal Amplification Control RNA | *0.2 μl | ||
| RNA | 3 µl |
* Amount varies with concentration of IAC RNA. The amount of IAC template needs to be adjusted based on the prepared stock concentration to report Cycle threshold at about 20-25 PCR cycles when no inhibition is present in the reaction.
+ Final concentration for 25 µl reaction is 4.0mM.
Table 6. MNV RT-qPCR Cycling Conditions: Cepheid Smart Cycler II
| Stage 1 | Stage 2 | Stage 3 | ||||||
|---|---|---|---|---|---|---|---|---|
| Hold | Hold | 3 Temperature Cycle | ||||||
| Temp | Sec | Optics | Temp | Sec | Optics | Repeat 45 times | ||
| 50 | 3000 | Off | 95 | 900 | Off | Temp | Sec | Optics |
| 95 | 15 | Off | ||||||
| 55 | 20 | Off | ||||||
| 62 | 60 | On | ||||||
Qualitative data analysis
On the SmartCycler II Instrument, set the following Analysis Settings for TxRed and Cy5 channels. Update analysis settings if they are changed before recording results.
- Usage: Assay
- Curve Analysis: Primary
- Threshold Setting: Manual
- Manual Threshold Fluorescence Units: 10.0
- Auto Min Cycle: 5
- Auto Max Cycle: 10
- Valid Min Cycle: 3
- Valid Max. Cycle: 60
- Background subtraction: ON
- Boxcar Avg. Cycles: 0
- Background Min. Cycle: 5
- Background Max. Cycle: 40
- Max Cycles 50
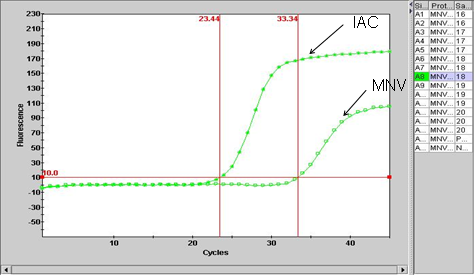
Primary fluorescence curves that cross the threshold will be recorded as "POS" and the cycle number when it crossed the threshold will be displayed in the Results Table view (Figure 3). Negative results are shown as "NEG". The TxRd and Cy5 channels correlate to IAC and the MNV targets, respectively. Results can also be viewed graphically. For example, Figure 3 is a graphical view for the two channels for the MNV and IAC.
Figure 3
Interpretation of real-time RT-PCR Results (Smart Cycler II)
Positive Samples:
Extraction controls are considered positive when value crosses threshold for Cy5 fluorescence channel for MNV spiked samples, IAC is positive within acceptable ranges, and positive MNV RT-qPCR control is positive and the negative RT-qPCR control is negative.
Negative samples:
Sample is "negative" when the MNV spiked control sample shows no detection for Cy5 fluorescence channel, the negative RT-qPCR control is negative, the positive MNV RT-qPCR control is positive for MNV, and the internal amplification control (IAC) is positive.
Invalid Samples:
- If the negative RT-qPCR control sample demonstrates positive results crossing the Cy5 threshold or if the IAC is negative, the RT-PCR assay must be repeated.
- The average of the Internal Amplification Control Ct values for the sample is more than 4 Ct s greater than the Negative Control Internal Amplification Control Ct value; the RT-PCR assay must be repeated using RNA from a newly extracted saved tube. If the repeat of the newly extracted sample yields average IAC Ct values greater than 4, the sample must be repeated from the beginning using additional food sample.
Note: for samples yielding continuous invalid results, analysts should contact subject matter expert (SME) for troubleshooting the protocol.
Detection of HAV in green onion.
The HAV multiplex assay is used to detect HAV in green onion RNA extracts. In figure 4, the graph demonstrates the TxRd and Cy5 channels correlating to the HAV and the IAC targets, respectively. Probable positive HAV samples or Cannot Rule Out (CRO) occurs when the primary fluorescence curve crosses the threshold for HAV and the IAC is positive. Sample data is considered invalid if: (1) the average of the HAV internal control Ct for the samples is ≥3.5 Ct values compared to the negative control and the MNV extraction control is not detected, and (2) false positive or false negative controls.
Figure 4.
Interpretation of real-time RT-PCR Results (Smart Cycler II)
Interpretation of Results:
For this HAV assay Cy5 is the HAV probe fluorescent label and that Texas Red (TxR) is the internal amplification control (IAC) probe fluorescent label.
Negative samples:
Sample is "negative" if the RT-qPCR negative control is negative for HAV, the RT-qPCR positive control is positive for HAV, the un-spiked sample (if included) is negative for HAV, the unknown is negative for HAV, and the internal amplification control (IAC) is positive. No further analysis is needed.
Positive Samples:
Sample is "positive" if the RT-qPCR negative control is negative for HAV, RT-qPCR positive control is positive for HAV, the unknown or spiked HAV RNA RT-PCR sample is positive for HAV, and the internal amplification control (IAC) is positive.
Invalid Samples:
- If the negative RT-qPCR control sample demonstrates positive results crossing the Cy5 threshold or if the IAC is negative, the RT-PCR assay must be repeated.
- The average of the Internal Amplification Control Ct values for the sample is more than 4 Ct’s greater than the Negative Control Internal Amplification Control Ct value, the RT-PCR assay must be repeated using RNA from a newly extracted saved tube. If the repeat of the newly extracted sample yields average IAC Ct values greater than 4, the sample must be repeated from the beginning using additional food sample.
Note: samples yielding continuous invalid results, analysts should contact subject matter expert (SME) for troubleshooting the protocol.
Probable positive HAV samples or Cannot Rule Out (CRO):
If the HAV sample is positive and the internal controls, negative controls, positive controls, and extraction controls are correct. A CRO protocol is available for use when required.
Appendices:
- Appendix A: Single Laboratory Validation Results
- Appendix B: Multi-laboratory Validation Results
- Appendix C: Multi-laboratory Concentration, Extraction, and Detection Results for HAV in Green Onion
- Appendix D: Buffer Solution and Extraction Reagent Recipes
- Appendix E: Inclusivity/Exclusivity Requirements for BAM Methods
- Appendix F: Template Preparation
- Appendix G: Murine Norovirus Extraction Control Preparation
References
- Arnal C, Ferre-Aubineau V, Mignotte B, Imbert-Marcille BM, Billaudel S. 1999. Quantification of hepatitis A virus in shellfish by competitive reverse transcription-PCR with coextraction of standard RNA. Appl. Environ. Microbiol. Jan; 65(1):322-6.
- DePaola, A., Jones, J.L., Woods, J., Burkhardt, W., III, Calci, K.R., Krantz, J.A., Bowers, J.C., Kasturi, K., Byars, R.H., Jacobs, E., Williams-Hill, D., & Nabe, K. 2010. Bacterial and viral pathogens in live oysters: 2007 United States market survey. Appl. Environ. Microbiol., 76, (9) 2754-2768.
- Dentinger CM, Bower WA, Nainan OV, Cotter SM, Myers G, Dubusky LM, Fowler S, Salehi ED, Bell BP. 2001. An outbreak of hepatitis A associated with green onions. J. Infect. Dis. Apr 15; 183(8):1273-6.
- European Food Safety Authority. 2012 Scientific Opinion of Norovirus (NoV) in oysters: methods, limits, and control options. EFSA Journal 10(1): 2500.
- Fiore AE. Hepatitis A transmitted by food. 2004. Clin. Infect. Dis. 2004 Mar 1; 38(5):705-15.
- Gardner SN, Kuczmarski TA, Vitalis EA, Slezak TR. 2003. Limitations of TaqMan PCR for detecting divergent viral pathogens illustrated by hepatitis A, B, C, and E viruses and human immunodeficiency virus. J. Clin. Microbiol. Jun; 41(6):2417-27
- Hewitt, J. Rivera-Aban, M., and Greening, G . E. 2009. Evaluation of murine norovirus as a surrogate for human norovirus and hepatitis A virus in heat inactivated studies. J. Appl. Microbiol. Jul; 107(1): 65-71.
- Hutin YJ, Pool V, Cramer EH, Nainan OV, Weth J, Williams IT, Goldstein ST, Gensheimer KF, Bell BP, Shapiro CN, Alter MJ, Margolis HS. 1999. A multistate, foodborne outbreak of hepatitis A. National Hepatitis A Investigation Team. N. Engl. J. Med. Feb 25; 340(8):595-602.
- Lednar, WM, Lemon, SM, Kilpatrick, JW, Redfield, RR, Fields, ML, Kelley, PW. 1985. Frequency of illness accociated with epidemic hepatitis A virus infection in adults. Aug; Am. J. Epidemiol., 122 (2): 226-33.
- Martil-Latil, S., Hennechart-Collette C, Guillier L, and Perelle, S. 2012. Comparison of tow extraction methods for the detection of hepatitis A in semi dried tomatoes and murine norovirus as a process control by duplex RT-qPCR. Food Microbiol. 31: 246-253.
- Niu MT, Polish LB, Robertson BH, Khanna BK, Woodruff BA, Shapiro CN, Miller MA, Smith JD, Gedrose JK, Alter MJ, et al. 1992. Multistate outbreak of hepatitis A associated with frozen strawberries. J. Infect. Dis. Sep;166(3):518-24.
- Pfaffl, MW. 2004. Quantification strategies in real-time PCR. A-Z of quantitative PCR. IUL. La Jolla, CA.
- Schwab KJ, Neill FH, Le Guyader F, Estes MK, Atmar RL. 2001. Development of a reverse transcription-PCR-DNA enzyme immunoassay for detection of "Norwalk-like" viruses and hepatitis A virus in stool and shellfish. Appl. Environ. Microbiol. Feb;67(2):742-9.
- Sair AI, D'Souza DH, Moe CL, Jaykus LA. 2002. Improved detection of human enteric viruses in foods by RT-PCR. J. Virol. Methods. Feb;100(1-2):57-69.
- Teunis, P.F., Moe, C.L., Liu, P., Miller, E., Lindesmith, L., Baric, R.S., Le, P.J., & Calderon, R.L. 2008. Norwalk virus: How infectious is it? J. Med. Virol., 80, (8) 1468-1476.