Veterinary Laboratory Investigation and Response Network
- Our Mission
- We Respond to Animal Illnesses Potentially Caused by Foods or Drugs
- Resources for Animal Owners and Veterinarians
- Tracking Antimicrobial Resistance in Bacteria from Sick Animals
- Promoting Antimicrobial Stewardship
- Vet-LIRN Laboratory Funding
- Ensuring Accurate Results
- Veterinary Student Opportunities
- Veterinarians, Want to Learn More?
- Preparing for and Responding to Emergencies
- COVID-19 Response
- Publications
Our Mission
To advance the CVM mission of protecting human and animal health by coordinating a network of veterinary diagnostic laboratories.
We Respond to Animal Illnesses Potentially Caused by Foods or Drugs
Is your animal sick? Do you think it was the food? Or a drug?
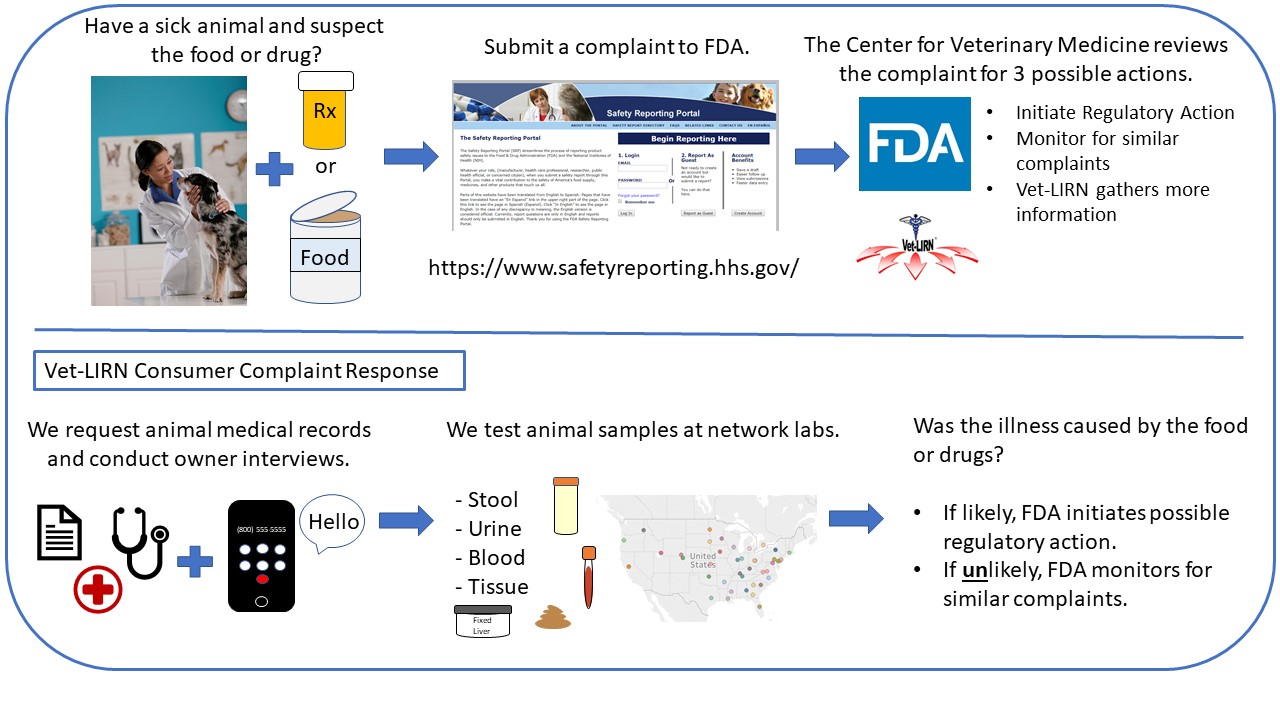
Figure 1. What Happens During a Consumer Complaint Response?
We respond to potential animal food issues, including performing non-regulatory testing (Figure 1).
We are an important part of the food safety team at CVM.
Learn more about some of our cases:
- Campylobacter Outbreak in Puppies
- Aflatoxin Recall
- Pig Ear Outbreak
- Dilated Cardiomyopathy in Some Cases of Pet Heart Disease
- Potentially Toxic Levels of Vitamin D in Several Dry Pet Foods
- The Melamine Story
- Jerky Pet Treats
Resources for Animal Owners and Veterinarians
- Safety Reporting Portal
- How to Report a Pet Food Complaint
- Reporting Problems with Horse or other Livestock Feed/Food
- Information for Veterinarians on Reporting Suspected Animal Food Issues
- Vet-LIRN Network Procedures for Veterinarians
- Vet-LIRN Network Procedures for Owners
- Vet-LIRN Network Procedures for Laboratories
- Vet-LIRN SARS-CoV-2 Supplemental Necropsy Sample Inventory Checklist
- Contact Vet-LIRN
- Pet Food Safety (CDC)
Tracking Antimicrobial Resistance in Bacteria from Sick Animals
Why track resistance in bacteria?
Antimicrobial resistance is an important public health issue because if bacteria become antibiotic-resistant, many infections will be more difficult to treat. In March of 2015, The first National Action Plan for Combating Antibiotic-Resistant Bacteria (CARB) was released to guide the government, public heath, healthcare, and veterinary partners in addressing antimicrobial resistance. In 2020, the second CARB plan was released. The new plan builds on the plan released in 2015 and presents coordinated, strategic actions that the United States Government will take in 2020-2025 by expanding evidence-based activities that have been shown to reduce antibiotic resistance. As part of this plan, Vet-LIRN was tasked to develop, expand, and maintain antimicrobial susceptibility testing (AST) and whole-genome sequencing (WGS) testing of veterinary pathogens isolated at veterinary diagnostic laboratories. To successfully monitor the antimicrobial susceptibility of bacterial pathogens, it is vital that veterinary diagnostic laboratories be incorporated into the nation’s other AMR monitoring activities. Vet-LIRN is committed to being a partner in this effort.
Vet-LIRN Antimicrobial Resistance Monitoring Program Background and Progress
- During 2017-2018, Vet-LIRN coordinated a two-year pilot project to evaluate the feasibility of using Vet-LIRN veterinary diagnostic laboratories to monitor the antimicrobial susceptibility of three veterinary pathogens: Escherichia coli and Staphylococcus pseudintermedius in dogs and Salmonella enterica in any animal host.
- Twenty Vet-LIRN Source diagnostic laboratories collected isolates and tested their antimicrobial susceptibility using Clinical and Laboratory Standards Institute (CLSI) methods.
- Approximately 5,000 isolates from clinically sick animals were collected and tested.
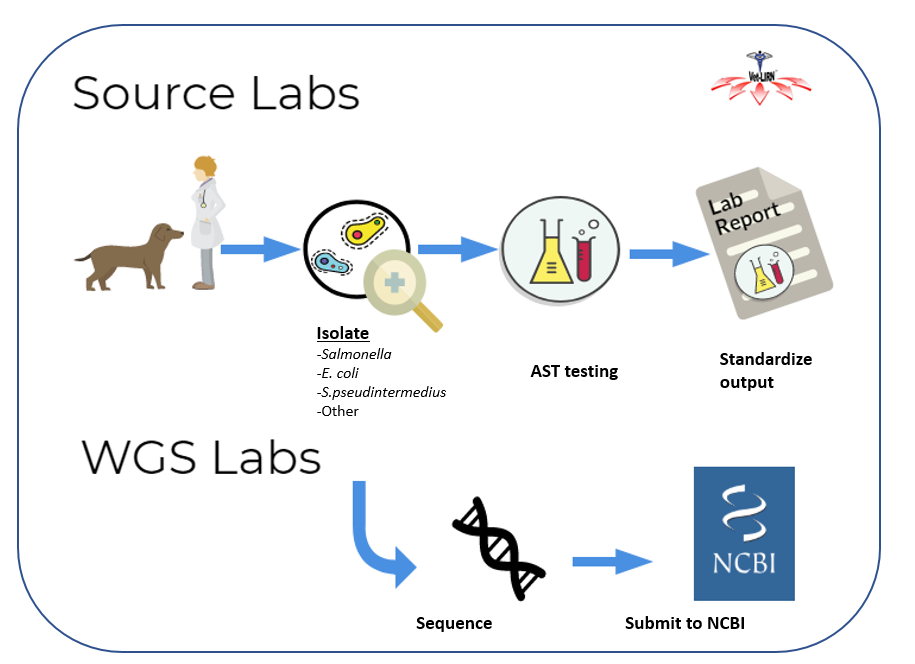
- WGS laboratories sequenced a subset of the isolates submitted by their Source labs and uploaded all sequences to National Center for Biotechnology Information (NCBI) through the GenomeTrakr program (Figure 2). Additional information about the pathogen (the organ it came from, the animal species, which part of the country) was reported. A publication summarizing the 2017 findings is available.
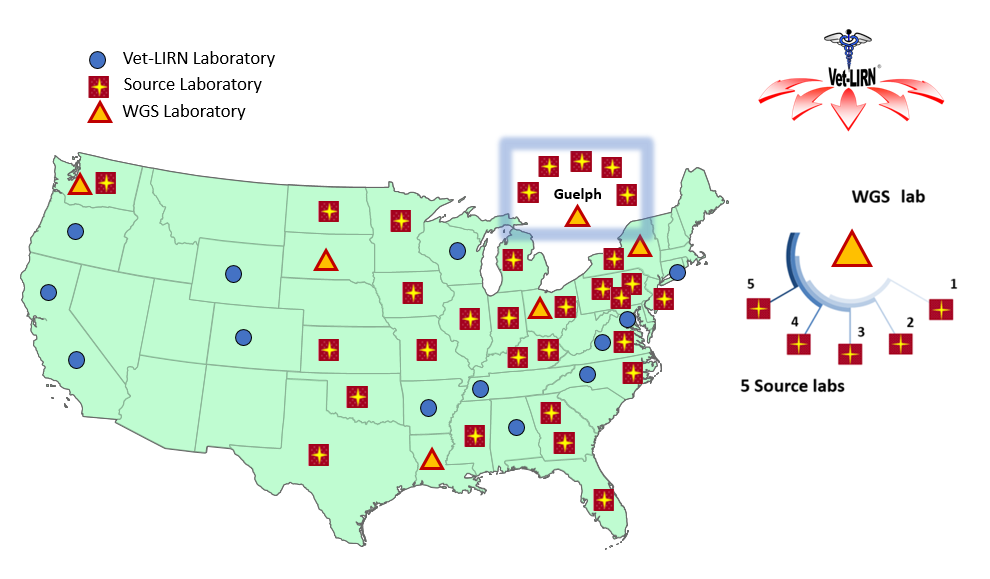
- In 2018-2019 additional labs began collecting and sequencing isolates. As of 2022, there are 30 Source laboratories collecting isolates (25 labs in U.S., and 5 labs in Canada) and six laboratories sequencing the isolates (Figure 3).
- As of January 2022, Vet-LIRN Source labs collected antimicrobial susceptibility data for more than 15,000 animal pathogen isolates and more than 4,000 isolates were sequenced.
- The data provides a snapshot of the susceptibility of pathogens being cultured at referral veterinary laboratories.
- Vet-LIRN partners with the National Antimicrobial Resistance Monitoring System (NARMS) to make the data public (2018 AMR data, 2019 AMR data). This animal pathogen data is reported in conjunction with the National Animal Health Laboratory Network (NAHLN).
Figure 2. Vet-LIRN AMR Monitoring: General Plan
Figure 3: Geographic distribution and organization of Vet-LIRN WGS and Source laboratories (2022)
Legend: Thirty Source laboratories (25 in the U.S. and 5 in Canada) (squares) are collecting isolates. Six WGS labs (triangles) each have 5 collaborating source labs each and sequence a subset of the isolates submitted by their source labs. Remaining Vet-LIRN laboratories not participating in the project are shown with circles.
Promoting Antimicrobial Stewardship
Along with tracking antimicrobial resistance, Vet-LIRN is working to promote antimicrobial stewardship in veterinary medicine. As described above, antimicrobial resistance is an important public health issue and use of antimicrobial drugs can contribute to the development of antimicrobial resistant bacteria. Antimicrobial stewardship involves using antimicrobials appropriately and only when necessary.
Vet-LIRN supports antimicrobial stewardship efforts by providing funding to veterinary colleges across the United States to work on several projects including:
- creating collaborative websites with antimicrobial resistance resources,
- generating veterinary hospital stewardship plans,
- developing educational materials for veterinary health professionals (including veterinarians and veterinary students), and
- developing educational materials for animal owners.
These materials consist of website content, fliers, videos, and other formats to encourage education and appropriate use of antimicrobials.
Vet-LIRN Laboratory Funding
Vet-LIRN Cooperative Agreements facilitate participation in Vet-LIRN activities such as consumer complaint response, emergency exercises, proficiency tests, and laboratory accreditation. The agreements also increase the agency’s capability to analyze an increased number of samples in the event of animal food- or drug-related illnesses or other large-scale emergency events that require increased testing of implicated diagnostic or animal food samples. Cooperative agreements allow network laboratories to request additional funds if they are participating in a specific Vet-LIRN project, such as the Antimicrobial Resistance (AMR) Project or if they are conducting whole-genome sequencing (WGS) work, or if their caseload is particularly heavy. Additional funds may also be provided to respond to emerging diseases such as COVID-19.
Ensuring Accurate Results
Vet-LIRN collaborates with the FDA’s Center for Food Safety and Nutrition (CFSAN) Division of Food Processing Science and Technology (Moffett Center) and the Institute for Food Safety and Health, Illinois Institute of Technology to conduct Proficiency Tests (PTs) and Interlaboratory Comparison Exercises (ICEs) to ensure FDA receives accurate test results from our network laboratories. Samples are sent to the laboratories and test results are submitted to Vet-LIRN. After data is evaluated, final reports are provided to the laboratories.
Recent Proficiency Tests and Inter-Laboratory Comparison Exercises
- Detecting SARS-COV-2
Timely to support veterinary diagnostic laboratories’ ability to evaluate the accuracy of their current testing methods.- Why is this important?
- From House Cats to Big Cats: How FDA Evaluated Methods for Detecting SARS-CoV-2 in Animals
- First Reported Cases of SARS-CoV-2 Infection in Companion Animals — New York, March–April 2020
- Pet Safety related to SARS-CoV-2 in Animals in the United States
- Evaluation for SARS-CoV-2 Testing in Animals
- Confirmed Cases of SARS-CoV-2 in Animals in the United States
- Detecting anticoagulant rodenticides in equine serum
- Why is this important? It evaluates network laboratories’ ability to identify the cause of an illness and act to rule out potential issues with animal foods or drugs.
- Evaluating Whole Genome Sequencing capabilities at network laboratories.
Timely due to confirmed usefulness of whole genome sequencing as a tool to help identify and limit the spread of foodborne illness outbreaks in FDA regulated foods.- Why is this important?
- Whole Genome Sequencing (WGS) Program
- FDA Assesses Impact of Whole Genome Sequencing Program
- Detecting Copper in bovine and canine liver
- Why is this important? It evaluates network laboratories’ ability to identify the cause of an illness and act to rule out potential issues with animal foods or drugs.
Veterinary Student Opportunities
Veterinarians are valuable partners in CVM’s mission to promote animal health. Vet-LIRN is committed to building relationships with the next generation of veterinary professionals. Veterinary students can apply for an externship through the FDA Veterinary Clerkship Program to train alongside Vet-LIRN team members. Students will learn more about CVM’s mission and be introduced to the many different roles that veterinarians play within the Center.
Veterinarians, Want to Learn More?
Vet-LIRN educates veterinarians and students about how to identify and report suspected animal food issues via webinars and case studies. Vet-LIRN speaks at various conferences, to veterinary interest groups, and to students. Please email [email protected] if you would like Vet-LIRN to speak to your organization.
Preparing for and Responding to Emergencies
Vet-LIRN participates in the planning, play, and evaluation of emergency preparedness and response activities. Such activities strengthen Vet-LIRN’s ability to establish and initiate strategies to coordinate the roles and responsibilities of veterinary diagnostics laboratories in real-world emergency events. Knowing the network laboratory capabilities and having routine interactions and exercises with the laboratories is key to any emergency response. Vet-LIRN routinely communicates with the following laboratory networks and programs to harmonize and leverage activities and participate in an integrated response to national emergencies:
- Integrated Consortium of Laboratory Networks (ICLN)
- National Animal Health Laboratory Network (NAHLN)
- The Food Emergency Response Network (FERN)
COVID-19 Response
Vet-LIRN is very active in supporting capacity and emergency response related to COVID-19. See publications associated with some of these activities in the publications section.
- In collaboration with numerous partners, Vet-LIRN offers an Inter-Laboratory Comparison Exercises (ICE) to evaluate SARS-CoV-2 detection assays at veterinary diagnostic laboratories, private industry, and other government partner laboratories. The first ICE (Round 1) was completed in October of 2020. In collaboration with the USDA’s National Animal Health Laboratory Network (NAHLN), FDA/CFSAN’s Moffett Center Proficiency Test Campus, Cornell University, United States Geological Survey, and the ICLN, Vet-LIRN conducted a second ICE (Round 2) to evaluate SARS-CoV-2 detection assays in June 2021.
- ICE1: Veterinary diagnostic laboratories developed tests for SARS-CoV-2 in animals in spring 2020. These methods were initially evaluated only in the originating laboratories. The Round 1 ICE allowed laboratories to evaluate their individual assays in comparison to assays run by other laboratories. Private commercial laboratories also participated. In August 2020, the Moffett Center shipped samples to over 40 participating laboratories. Sample preparation and analysis of results were completed following ISO Guidelines 13528, 16140, and 17043. Results showed that for RNA in buffer, 100% of samples were detected, and for samples requiring extraction, PCR methods provided almost perfect results.
- ICE 2: The round 2 ICE was conducted in June 2021 and included SARS-CoV-2 and non-SARS-CoV-2 coronavirus samples at various concentrations to evaluate sensitivity and specificity of participants’ methods and their ability to detect emerging variants. Participant laboratories included those in the Vet-LIRN and NAHLN networks, as well as CDC, DOD, USGS, EPA, and private laboratories. ICE2 is even more relevant now because veterinary diagnostic laboratories are testing human samples for the SARS-CoV-2 virus under Clinical Laboratory Improvement Amendments (CLIA) certifications and have tested millions of human diagnostic samples.
- Vet-LIRN, in collaboration with FDA’s Center for Veterinary Medicine’s Office of Surveillance and Compliance, facilitates necropsies of animals, excluding production animals, that tested positive for or were exposed to SARS-CoV-2. Vet-LIRN partnered with USDA’s National Veterinary Services Laboratory, Department of Defense, and CDC to develop a sample checklist to standardize sample collection and archiving for partners conducting necropsies. This work is a part of a One Health approach that includes a collaboration with the CDC, state and local veterinarians, and Vet-LIRN to allow network laboratories to conduct necropsies on animals across the nation. To date, three cats, one dog, and one tiger were necropsied through this collaborative process. This work is important because we are still learning about the disease process in natural exposures in animals. In addition, this collaboration builds on the One Health framework.
- Vet-LIRN advances animal diagnostics for COVID-19 with various grant projects for network laboratories, including those that increase testing capacity, promote variant detection, and result in the development and validation of new diagnostic methods. This work reinforces the comprehensive One Health approach as part of the pandemic response.
While Vet-LIRN’s efforts related to COVID-19 are focused on emergency response, they also promote and protect human and animal health. The veterinary diagnostic laboratory community is building laboratory capacity, training scientists, and providing critical scientific information to federal stakeholders. Vet-LIRN is focused on ensuring results gathered by network laboratories are accurate and meaningful to advance our ability to respond to the pandemic.
Publications (Listed past 5 years)
2022
Rotstein, D. S., Peloquin, S., Proia, K., Hart, E., Lee, J., Vyhnal, K. K., . . . Ghai, R. (2022). Investigation of SARS-CoV-2 infection and associated lesions in exotic and companion animals. Vet Pathol, 3009858211067467. doi:10.1177/03009858211067467
Mitchell, P. K., Wang, L., Stanhope, B. J., Cronk, B. D., Anderson, R., Mohan, S., . . . Goodman, L. B. (2022). Multi-laboratory evaluation of the Illumina iSeq platform for whole genome sequencing of Salmonella, Escherichia coli and Listeria. Microb Genom, 8(2). doi:10.1099/mgen.0.000717
2021
Deng, K., Uhlig, S., Ip, H. S., Lea Killian, M., Goodman, L. B., Nemser, S., . . . Reimschuessel, R. (2021). Interlaboratory comparison of SARS-CoV2 molecular detection assays in use by U.S. veterinary diagnostic laboratories. J Vet Diagn Invest, 33(6), 1039-1051. doi:10.1177/10406387211029913
Girard, L., Herath, K., Escobar, H., Reimschuessel, R., Ceric, O., & Jayasuriya, H. (2021). Development of UHPLC/Q-TOF Analysis Method to Screen Glycerin for Direct Detection of Process Contaminants 3-Monochloropropane-1,2-diol Esters (3-MCPDEs) and Glycidyl Esters (GEs). Molecules, 26(9). doi:10.3390/molecules26092449
Nemser, S., Lindemann, S., Chen, Y., Lopez, S., Pickens, S., Ulaszek, J., . . . Reddy, R. (2021). A review of proficiency exercises offered by the Veterinary Laboratory Investigation and Response Network (Vet-LIRN) and Moffett Proficiency Testing Laboratory from 2012 to 2018. Accreditation and Quality Assurance, 26(3), 143-156. doi:10.1007/s00769-021-01471-x
Rotstein, D., Jones, J. L., Buchweitz, J., Refsal, K. R., Wilson, R., Yanes, E. G., . . . Reimschuessel, R. (2021). Pet Food-Associated Dietary Exogenous Thyrotoxicosis: Retrospective Study (2016-2018) and Clinical Considerations. Top Companion Anim Med, 43, 100521. doi:10.1016/j.tcam.2021.100521
Peloquin, S. K., Rotstein, D. S., Jones, J. L., Guag, J., Carey, L., Palmer, L. A., . . . Reimschuessel, R. (2021). Presumed Choline Chloride Toxicosis in Cats With Positive Ethylene Glycol Tests After Consuming a Recalled Cat Food. Top Companion Anim Med, 44, 100548. doi:10.1016/j.tcam.2021.100548
Taghvaei, M., Tonyali, B., Sommers, C., Ceric, O., Linghu, Z., Smith, J. S., & Yucel, U. (2021). Formation kinetics of radiolytic lipid products in model food–lipid systems with gamma irradiation. Journal of the American Oil Chemists' Society, 98(7), 737-746. doi:https://doi.org/10.1002/aocs.12513
Tkachenko, A., Benson, K., Mostrom, M., Guag, J., Reimschuessel, R., & Webb, B. (2021). Extensive evaluation via blinded testing of an UHPLC-MS/MS method for quantitation of ten ergot alkaloids in rye and wheat grains. J AOAC Int. doi:10.1093/jaoacint/qsaa173
Tyson, G. H., Ceric, O., Guag, J., Nemser, S., Borenstein, S., Slavic, D., . . . Reimschuessel, R. (2021). Genomics accurately predicts antimicrobial resistance in Staphylococcus pseudintermedius collected as part of Vet-LIRN resistance monitoring. Veterinary Microbiology, 254, 109006. doi:https://doi.org/10.1016/j.vetmic.2021.109006
Vudathala, D., Cummings, M., Tkachenko, A., Guag, J., Reimschuessel, R., & Murphy, L. (2021). A Lateral Flow Method for Aflatoxin B1 in Dry Dog Food: An Inter-Laboratory Trial. J AOAC Int. doi:10.1093/jaoacint/qsaa175
2020
Cole, S. D., Peak, L., Tyson, G. H., Reimschuessel, R., Ceric, O., & Rankin, S. C. (2020). New Delhi Metallo-β-Lactamase-5-Producing Escherichia coli in Companion Animals, United States. Emerg Infect Dis, 26(2), 381-383. doi:10.3201/eid2602.191221
Nichols, M., Stevenson, L., Koski, L., Basler, C., Wise, M., Whitlock, L., . . . Williams, I. T. (2020). Detecting national human enteric disease outbreaks linked to animal contact in the United States of America. Rev Sci Tech, 39(2), 471-480. doi:10.20506/rst.39.2.3098
Taghvaei, M., Sommers, C., Ceric, O., Hussain, F., Yucel, U., & Smith, J. S. (2020). Solid-phase micro extraction of food irradiation marker 2-dodecylcyclobutanone (2-DCB) from chicken jerky treated with glycerol. J Food Sci, 85(8), 2608-2614. doi:10.1111/1750-3841.15322
Tonyali, B., Sommers, C., Ceric, O., Smith, J. S., & Yucel, U. (2020). An analysis of cellulose- and dextrose-based radicals in sweet potatoes as irradiation markers. J Food Sci, 85(9), 2745-2753. doi:10.1111/1750-3841.15359
Vudathala, D., Klobut, J., Cummings, M., Tkachenko, A., Reimschuessel, R., & Murphy, L. (2020). Multilaboratory Evaluation of a Lateral Flow Method for Aflatoxin B1 Analysis in Dry Dog Food. J AOAC Int, 103(2), 480-488. doi:10.5740/jaoacint.19-0020
2019
Brill, R. W., Horodysky, A. Z., Place, A. R., Larkin, M. E. M., & Reimschuessel, R. (2019). Effects of dietary taurine level on visual function in European sea bass (Dicentrarchus labrax). PLoS One, 14(6), e0214347. doi:10.1371/journal.pone.0214347
Ceric, O., Tyson, G. H., Goodman, L. B., Mitchell, P. K., Zhang, Y., Prarat, M., . . . Reimschuessel, R. J. B. V. R. (2019). Enhancing the one health initiative by using whole genome sequencing to monitor antimicrobial resistance of animal pathogens: Vet-LIRN collaborative project with veterinary diagnostic laboratories in United States and Canada. 15(1), 130. doi:10.1186/s12917-019-1864-2
Du, X., Schrunk, D. E., Imerman, P. M., Smith, L., Francis, K., Tahara, J., . . . Rumbeiha, W. K. (2019). Evaluation of a Diagnostic Method to Quantify Aflatoxins B(1) and M(1) in Animal Liver by High-Performance Liquid Chromatography with Fluorescence Detection. J AOAC Int, 102(5), 1530-1534. doi:10.5740/jaoacint.18-0355
Jones, J. L., Wang, L., Ceric, O., Nemser, S. M., Rotstein, D. S., Jurkovic, D. A., . . . Reimschuessel, R. (2019). Whole genome sequencing confirms source of pathogens associated with bacterial foodborne illness in pets fed raw pet food. J Vet Diagn Invest, 1040638718823046. doi:10.1177/1040638718823046
Tyson, G. H., Li, C., Ceric, O., Reimschuessel, R., Cole, S., Peak, L., & Rankin, S. C. (2019). Complete Genome Sequence of a Carbapenem-Resistant Escherichia coli Isolate with bla NDM-5 from a Dog in the United States. Microbiol Resour Announc, 8(34). doi:10.1128/MRA.00872-19
2018
Buchweitz, J. P., Johnson, M., Jones, J. L., & Lehner, A. F. (2018). Development of a Quantitative Gas Chromatography-Tandem Mass Spectrometry Method for the Determination of Pentobarbital in Dog Food. J Agric Food Chem, 66(42), 11166-11169. doi:10.1021/acs.jafc.8b04178
Harrison, L. M., Gaines, D. W., Babu, U. S., Balan, K. V., Reimschuessel, R., Do, A. B., . . . Williams, K. M. (2018). Diet-induced obesity precipitates kidney dysfunction and alters inflammatory mediators in mice treated with Shiga Toxin 2. Microb Pathog, 123, 250-258. doi:10.1016/j.micpath.2018.07.015
Jones, J. L., Rotstein, D. S., Ceric, O., Nemser, S. M., & Reimschuessel, R. (2018). Information for veterinarians on reporting suspected animal food issues. J Am Vet Med Assoc, 253(5), 550-553. doi:10.2460/javma.253.5.550
2017
Mitchell, E. P., Church, M. E., Nemser, S. M., Yakes, B. J., Evans, E. R., Reimschuessel, R., . . . Terio, K. A. (2017). Pathology and Epidemiology of Oxalate Nephrosis in Cheetahs. Veterinary Pathology, 54(6), 977-985. doi:10.1177/0300985817728556
Reimschuessel, R., Grabenstein, M., Guag, J., Nemser, S. M., Song, K., Qiu, J., . . . Okwumabua, O. (2017). Multilaboratory Survey To Evaluate Salmonella Prevalence in Diarrheic and Nondiarrheic Dogs and Cats in the United States between 2012 and 2014. J Clin Microbiol, 55(5), 1350-1368. doi:10.1128/jcm.02137-16
Smith, L. L., Liang, B., Booth, M. C., Filigenzi, M. S., Tkachenko, A., & Gaskill, C. L. (2017). Development and Validation of Quantitative Ultraperformance Liquid Chromatography–Tandem Mass Spectrometry Assay for Anticoagulant Rodenticides in Liver. Journal of Agricultural and Food Chemistry, 65(31), 6682-6691. doi:10.1021/acs.jafc.7b02280
Publications (Vet-LIRN funded) (Listed past 3 years)
2022
Zehr, J. D., Pond, S. L. K., Martin, D. P., Ceres, K., Whittaker, G. R., Millet, J. K., . . . Stanhope, M. J. (2022). Recent Zoonotic Spillover and Tropism Shift of a Canine Coronavirus Is Associated with Relaxed Selection and Putative Loss of Function in NTD Subdomain of Spike Protein. Viruses, 14(5). doi:10.3390/v14050853
2021
Gioia, G., Addis, M. F., Goodman, L. B., Mitchell, P. K., Thompson, B., Goodrich, E., & Moroni, P. (2021). Draft Genome Sequence of Acholeplasma laidlawii Isolated from the Conjunctiva of a Heifer with Infectious Bovine Keratoconjunctivitis. Microbiol Resour Announc, 10(4). doi:10.1128/mra.01345-20
Oh, C., Sashittal, P., Zhou, A., Wang, L., El-Kebir, M., & Nguyen, T. H. (2021). Regional and temporal variations affect the accuracy of variant-specific SARS-CoV-2 PCR assays. medRxiv, 2021.2011.2008.21266083. doi:10.1101/2021.11.08.21266083
Podico, G., Gray, S. M., Wang, L., & Canisso, I. F. (2021). A novel Streptococcus species causing clinical mastitis in a pregnant donkey. J Vet Diagn Invest, 33(5), 979-983. doi:10.1177/10406387211027306
Savard, C., Ariel, O., Fredrickson, R., Wang, L., & Broes, A. (2021). Detection and genome characterization of bovine kobuvirus (BKV) in faecal samples from diarrhoeic calves in Quebec, Canada. Transbound Emerg Dis. doi:10.1111/tbed.14086
Savard, C., Provost, C., Ariel, O., Morin, S., Fredrickson, R., Gagnon, C. A., . . . Wang, L. (2021). First report and genomic characterization of a bovine-like coronavirus causing enteric infection in an odd-toed non-ruminant species (Indonesian tapir, Acrocodia indica) during an outbreak of winter dysentery in a zoo. Transbound Emerg Dis. doi:10.1111/tbed.14300
2020
Brooks, Y. M., Spirito, C. M., Bae, J. S., Hong, A., Mosier, E. M., Sausele, D. J., . . . Richardson, R. E. (2020). Fecal indicator bacteria, fecal source tracking markers, and pathogens detected in two Hudson River tributaries. Water Res, 171, 115342. doi:10.1016/j.watres.2019.115342
Cummings, K. J., Mitchell, P. K., Rodriguez-Rivera, L. D., & Goodman, L. B. (2020). Sequence analysis of Salmonella enterica isolates obtained from shelter dogs throughout Texas. Vet Med Sci, 6(4), 975-979. doi:10.1002/vms3.320
McGeehan, S., Baszler, T., Gaskill, C., Johnson, J., Smith, L., Raisbeck, M., . . . Talcott, P. (2020). Interlaboratory comparison of heavy metal testing in animal diagnostic specimens and feed using inductively coupled plasma-mass spectrometry. J Vet Diagn Invest, 1040638720903115. doi:10.1177/1040638720903115
Stout, A. E., Hofmar-Glennon, H. G., André, N. M., Goodman, L. B., Anderson, R. R., Mitchell, P. K., . . . Goodrich, E. L. (2021). Infectious disease surveillance of apparently healthy horses at a multi-day show using a novel nanoscale real-time PCR panel. J Vet Diagn Invest, 33(1), 80-86. doi:10.1177/1040638720972096
Wang, L., Mitchell, P. K., Calle, P. P., Bartlett, S. L., McAloose, D., Killian, M. L., . . . Torchetti, M. K. (2020). Complete Genome Sequence of SARS-CoV-2 in a Tiger from a U.S. Zoological Collection. Microbiol Resour Announc, 9(22). doi:10.1128/mra.00468-20