alveoli
How Severe COVID-19 Can Tragically Lead to Lung Failure and Death
Posted on by Dr. Francis Collins

More than 3 million people around the world, now tragically including thousands every day in India, have lost their lives to severe COVID-19. Though incredible progress has been made in a little more than a year to develop effective vaccines, diagnostic tests, and treatments, there’s still much we don’t know about what precisely happens in the lungs and other parts of the body that leads to lethal outcomes.
Two recent studies in the journal Nature provide some of the most-detailed analyses yet about the effects on the human body of SARS-CoV-2, the coronavirus that causes COVID-19 [1,2]. The research shows that in people with advanced infections, SARS-CoV-2 often unleashes a devastating series of host events in the lungs prior to death. These events include runaway inflammation and rampant tissue destruction that the lungs cannot repair.
Both studies were supported by NIH. One comes from a team led by Benjamin Izar, Columbia University, New York. The other involves a group led by Aviv Regev, now at Genentech, and formerly at Broad Institute of MIT and Harvard, Cambridge, MA.
Each team analyzed samples of essential tissues gathered from COVID-19 patients shortly after their deaths. Izar’s team set up a rapid autopsy program to collect and freeze samples within hours of death. He and his team performed single-cell RNA sequencing on about 116,000 cells from the lung tissue of 19 men and women. Similarly, Regev’s team developed an autopsy biobank that included 420 total samples from 11 organ systems, which were used to generate multiple single-cell atlases of tissues from the lung, kidney, liver, and heart.
Izar’s team found that the lungs of people who died of COVID-19 were filled with immune cells called macrophages. While macrophages normally help to fight an infectious virus, they seemed in this case to produce a vicious cycle of severe inflammation that further damaged lung tissue. The researchers also discovered that the macrophages produced high levels of IL-1β, a type of small inflammatory protein called a cytokine. This suggests that drugs to reduce effects of IL-1β might have promise to control lung inflammation in the sickest patients.
As a person clears and recovers from a typical respiratory infection, such as the flu, the lung repairs the damage. But in severe COVID-19, both studies suggest this isn’t always possible. Not only does SARS-CoV-2 destroy cells within air sacs, called alveoli, that are essential for the exchange of oxygen and carbon dioxide, but the unchecked inflammation apparently also impairs remaining cells from repairing the damage. In fact, the lungs’ regenerative cells are suspended in a kind of reparative limbo, unable to complete the last steps needed to replace healthy alveolar tissue.
In both studies, the lung tissue also contained an unusually large number of fibroblast cells. Izar’s team went a step further to show increased numbers of a specific type of pathological fibroblast, which likely drives the rapid lung scarring (pulmonary fibrosis) seen in severe COVID-19. The findings point to specific fibroblast proteins that may serve as drug targets to block deleterious effects.
Regev’s team also describes how the virus affects other parts of the body. One surprising discovery was there was scant evidence of direct SARS-CoV-2 infection in the liver, kidney, or heart tissue of the deceased. Yet, a closer look heart tissue revealed widespread damage, documenting that many different coronary cell types had altered their genetic programs. It’s still to be determined if that’s because the virus had already been cleared from the heart prior to death. Alternatively, the heart damage might not be caused directly by SARS-CoV-2, and may arise from secondary immune and/or metabolic disruptions.
Together, these two studies provide clearer pictures of the pathology in the most severe and lethal cases of COVID-19. The data from these cell atlases has been made freely available for other researchers around the world to explore and analyze. The hope is that these vast data sets, together with future analyses and studies of people who’ve tragically lost their lives to this pandemic, will improve our understanding of long-term complications in patients who’ve survived. They also will now serve as an important foundational resource for the development of promising therapies, with the goal of preventing future complications and deaths due to COVID-19.
References:
[1] A molecular single-cell lung atlas of lethal COVID-19. Melms JC, Biermann J, Huang H, Wang Y, Nair A, Tagore S, Katsyv I, Rendeiro AF, Amin AD, Schapiro D, Frangieh CJ, Luoma AM, Filliol A, Fang Y, Ravichandran H, Clausi MG, Alba GA, Rogava M, Chen SW, Ho P, Montoro DT, Kornberg AE, Han AS, Bakhoum MF, Anandasabapathy N, Suárez-Fariñas M, Bakhoum SF, Bram Y, Borczuk A, Guo XV, Lefkowitch JH, Marboe C, Lagana SM, Del Portillo A, Zorn E, Markowitz GS, Schwabe RF, Schwartz RE, Elemento O, Saqi A, Hibshoosh H, Que J, Izar B. Nature. 2021 Apr 29.
[2] COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Delorey TM, Ziegler CGK, Heimberg G, Normand R, Shalek AK, Villani AC, Rozenblatt-Rosen O, Regev A. et al. Nature. 2021 Apr 29.
Links:
COVID-19 Research (NIH)
Izar Lab (Columbia University, New York)
Aviv Regev (Genentech, South San Francisco, CA)
NIH Support: National Center for Advancing Translational Sciences; National Heart, Lung, and Blood Institute; National Cancer Institute; National Institute of Allergy and Infectious Diseases; National Institute of Diabetes and Digestive and Kidney Diseases; National Human Genome Research Institute; National Institute of Mental Health; National Institute on Alcohol Abuse and Alcoholism
Mapping Severe COVID-19 in the Lungs at Single-Cell Resolution
Posted on by Dr. Francis Collins
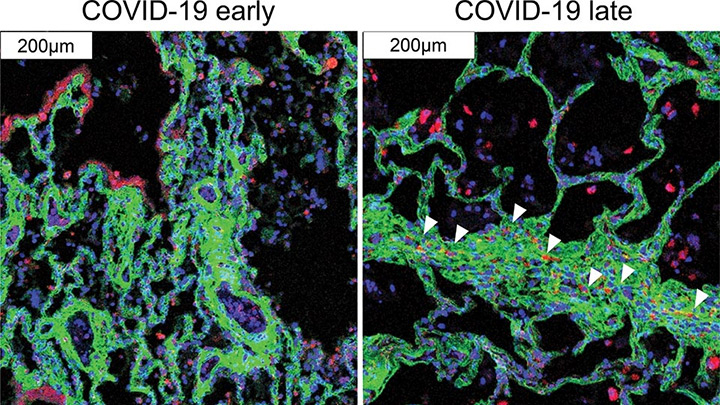
A crucial question for COVID-19 researchers is what causes progression of the initial infection, leading to life-threatening respiratory illness. A good place to look for clues is in the lungs of those COVID-19 patients who’ve tragically lost their lives to acute respiratory distress syndrome (ARDS), in which fluid and cellular infiltrates build up in the lung’s air sacs, called alveoli, keeping them from exchanging oxygen with the bloodstream.
As shown above, a team of NIH-funded researchers has done just that, capturing changes in the lungs over the course of a COVID-19 infection at unprecedented, single-cell resolution. These imaging data add evidence that SARS-CoV-2, the coronavirus that causes COVID-19, primarily infects cells at the surface of the air sacs. Their findings also offer valuable clues for treating the most severe consequences of COVID-19, suggesting that a certain type of scavenging immune cell might be driving the widespread lung inflammation that leads to ARDS.
The findings, published in Nature [1], come from Olivier Elemento and Robert E. Schwartz, Weill Cornell Medicine, New York. They already knew from earlier COVID-19 studies about the body’s own immune response causing the lung inflammation that leads to ARDS. What was missing was an understanding of the precise interplay between immune cells and lung tissue infected with SARS-CoV-2. It also wasn’t clear how the ARDS seen with COVID-19 compared to the ARDS seen in other serious respiratory diseases, including influenza and bacterial pneumonia.
Traditional tissue analysis uses chemical stains or tagged antibodies to label certain proteins and visualize important features in autopsied human tissues. But using these older techniques, it isn’t possible to capture more than a few such proteins at once. To get a more finely detailed view, the researchers used a more advanced technology called imaging mass cytometry [2].
This approach uses a collection of lanthanide metal-tagged antibodies to label simultaneously dozens of molecular markers on cells within tissues. Next, a special laser scans the labeled tissue sections, which vaporizes the heavy metal tags. As the metals are vaporized, their distinct signatures are detected in a mass spectrometer along with their spatial position relative to the laser. The technique makes it possible to map precisely where a diversity of distinct cell types is located in a tissue sample with respect to one another.
In the new study, the researchers applied the method to 19 lung tissue samples from patients who had died of severe COVID-19, acute bacterial pneumonia, or bacterial or influenza-related ARDS. They included 36 markers to differentiate various types of lung and immune cells as well as the SARS-CoV-2 spike protein and molecular signs of immune activation, inflammation, and cell death. For comparison, they also mapped four lung tissue samples from people who had died without lung disease.
Altogether, they captured more than 200 lung tissue maps, representing more than 660,000 cells across all the tissues sampled. Those images showed in all cases that respiratory infection led to a thickening of the walls surrounding alveoli as immune cells entered. They also showed an increase in cell death in infected compared to healthy lungs.
Their maps suggest that what happens in the lungs of COVID-19 patients who die with ARDS isn’t entirely unique. It’s similar to what happens in the lungs of those with other life-threatening respiratory infections who also die with ARDS.
They did, however, reveal a potentially prominent role in COVID-19 for white blood cells called macrophages. The results showed that macrophages are much more abundant in the lungs of severe COVID-19 patients compared to other lung infections.
In late COVID-19, macrophages also increase in the walls of alveoli, where they interact with lung cells known as fibroblasts. This suggests these interactions may play a role in the buildup of damaging fibrous tissue, or scarring, in the alveoli that tends to be seen in severe COVID-19 respiratory infections.
While the virus initiates this life-threatening damage, its progression may not depend on the persistence of the virus, but on an overreaction of the immune system. This may explain why immunomodulatory treatments like dexamethasone can provide benefit to the sickest patients with COVID-19. To learn even more, the researchers are making their data and maps available as a resource for scientists around the world who are busily working to understand this devastating illness and help put an end to the terrible toll caused by this pandemic.
References:
[1] The spatial landscape of lung pathology during COVID-19 progression. Rendeiro AF, Ravichandran H, Bram Y, Chandar V, Kim J, Meydan C, Park J, Foox J, Hether T, Warren S, Kim Y, Reeves J, Salvatore S, Mason CE, Swanson EC, Borczuk AC, Elemento O, Schwartz RE. Nature. 2021 Mar 29.
[2] Mass cytometry imaging for the study of human diseases-applications and data analysis strategies. Baharlou H, Canete NP, Cunningham AL, Harman AN, Patrick E. Front Immunol. 2019 Nov 14;10:2657.
Links:
COVID-19 Research (NIH)
Elemento Lab (Weill Cornell Medicine, New York)
Schwartz Lab (Weill Cornell Medicine)
NIH Support: National Center for Advancing Translational Sciences; National Institute of Allergy and Infectious Diseases; National Institute of Diabetes and Digestive and Kidney Diseases; National Cancer Institute
Mini-Lungs in a Lab Dish Mimic Early COVID-19 Infection
Posted on by Dr. Francis Collins

Researchers have become skilled at growing an array of miniature human organs in the lab. Such lab-grown “organoids” have been put to work to better understand diabetes, fatty liver disease, color vision, and much more. Now, NIH-funded researchers have applied this remarkable lab tool to produce mini-lungs to study SARS-CoV-2, the coronavirus that causes COVID-19.
The intriguing bubble-like structures (red/clear) in the mini-lung pictured above represent developing alveoli, the tiny air sacs in our lungs, where COVID-19 infections often begin. In this organoid, the air sacs consist of many thousands of cells, all of which arose from a single adult stem cell isolated from tissues found deep within healthy human lungs. When carefully nurtured in lab dishes, those so-called alveolar epithelial type-2 cells (AT2s) begin to multiply. As they grow, they spontaneously assemble into structures that closely resemble alveoli.
A team led by Purushothama Rao Tata, Duke University School of Medicine, Durham, NC, developed these mini-lungs in a quest to understand how adult stem cells help to regenerate damaged tissue in the deepest recesses of the lungs, where SARS-CoV-2 attacks. In earlier studies, the researchers had shown it was possible for these cells to produce miniature alveoli. But there was a problem: the “soup” they used to nurture the growing cells included ingredients that weren’t well defined, making it hard to characterize the experiments fully.
In the study, now reported in Cell Stem Cell, the researchers found a way to simplify and define that brew. For the first time, they could produce mini-lungs consisting only of human lung cells. By growing them in large numbers in the lab, they can now learn more about SARS-CoV-2 infection and look for new ways to prevent or treat it.
Tata and his collaborators at the University of North Carolina, Chapel Hill, have already confirmed that SARS-CoV-2 infects the mini-lungs via the critical ACE2 receptor, just as the virus is known to do in the lungs of an infected person.
Interestingly, the cells also produce cytokines, inflammatory molecules that have been tied to tissue damage. The findings suggest the cytokine signals may come from the lungs themselves, even before immune cells arrive on the scene.
The heavily infected lung cells eventually self-destruct and die. In an unexpected turn of events, they even induce cell death in some neighboring healthy cells that are not infected. The relevance of the studies to the clinic was boosted by the finding that the gene activity patterns in the mini-lungs are a close match to those found in samples taken from six patients with severe COVID-19.
Now that he’s got the recipe down, Tata is busy making organoids and helping to model COVID-19 infections, with the hope of identifying and testing promising new treatments. It’s clear these mini-lungs are breathing some added life into the basic study of COVID-19.
Reference:
[1] Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2-mediated interferon responses and pneumocyte dysfunction. Katsura H, Sontake V, Tata A, Kobayashi Y, Edwards CE, Heaton BE, Konkimalla A, Asakura T, Mikami Y, Fritch EJ, Lee PJ, Heaton NS, Boucher RC, Randell SH, Baric RS, Tata PR. Cell Stem Cell. 2020 Oct 21:S1934-5909(20)30499-9.
Links:
Coronavirus (COVID-19) (NIH)
Tata Lab (Duke University School of Medicine, Durham, NC)
NIH Support: National Institute of Allergy and Infectious Diseases; National Heart, Lung, and Blood Institute; National Institute of General Medical Sciences; National Institute of Diabetes and Digestive and Kidney Diseases
Progress Toward 3D Printed Human Organs
Posted on by Dr. Francis Collins
There’s considerable excitement that 3D printing technology might one day allow scientists to produce fully functional replacement organs from one’s own cells. While there’s still a lot to learn, this video shows just some of the amazing progress that’s now being made.
The video comes from a bioengineering team at Rice University, Houston, that has learned to bioprint the small air sacs in the lungs. When hooked up to a machine that pulsed air in and out of the air sacs, the rhythmic movement helped to mix red blood cells traveling through an associated blood vessel network. Those red cells also took up oxygen in much the way that blood vessels do when surrounding the hundreds of millions of air sacs in our lungs.
As mentioned in the video, one of the biggest technical hurdles in growing fully functional replacement tissues and organs is to find a way to feed the growing tissues with a blood supply and to remove waste products. In this study recently published in Science [1], the NIH-supported team cleared this hurdle by creating an open-source bioprinting technology they call SLATE, which is short for “stereo-lithography apparatus for tissue engineering.”
The SLATE system “grows” soft hydrogel scaffolds one layer at a time. Each layer is printed using a liquid pre-hydrogel solution that solidifies when exposed to blue light. By also projecting light into the hydrogel as a pixelated 3D shape, it’s possible to print complex 3D structures within minutes.
When the researchers first started, their printouts lacked the high resolution, submillimeter-scale channels needed to generate intricate vascular networks. In other manufacturing arenas, light-absorbing chemicals have helped control the conversion from liquid to solid in a very fine polymer layer. But these industrial light-absorbing chemicals are highly toxic and therefore unsuitable for scaffolds that grow living tissues and organs.
The researchers, including Bagrat Grigoryan, Jordan Miller, and Kelly Stevens, wondered whether they could swap out those noxious ingredients with synthetic and natural food dyes widely used in the food industry. These dyes include curcumin, anthocyanin, and tartrazine (yellow dye #5). Their studies showed that those fully biocompatible dyes worked as effective light absorbers, allowing the scientists to recreate the complex architectures of human vasculature. Importantly, the living cells survived within the soft scaffold!
These models are already yielding intriguing new insights into the vascular structures found within our organs and how those architectures may influence function in ways that hadn’t been well understood. In the near term, tissues and organs grown on such scaffolds might also find use as sophisticated, 3D tissue “chips,” with potential for use in studies to predict whether drugs will be safe in humans.
In the long term, this technology may allow production of replacement organs from those needing them. More than 100,000 men, women, and children are on the national transplant waiting list in the United States alone and 20 people die each day waiting for a transplant [2]. Ultimately, with the aid of bioprinting advances like this one, perhaps one day we’ll have a ready supply of perfectly matched and fully functional organs.
References:
[1] Multivascular networks and functional intravascular topologies within biocompatible hydrogels. Grigoryan B, Paulsen SJ, Corbett DC, Sazer DW, Fortin CL, Zaita AJ, Greenfield PT, Calafat NJ, Gounley JP, Ta AH, Johansson F, Randles A, Rosenkrantz JE, Louis-Rosenberg JD, Galie PA, Stevens KR, Miller JS. Science. 2019 May 3;364(6439):458-464.
[2] Organ Donor Statistics, Health Resources & Services Administration, October 2018.
Links:
Tissue Engineering and Regenerative Medicine (National Institute of Biomedical Imaging and Bioengineering/NIH)
Tissue Chip for Drug Screening (National Center for Advancing Translational Sciences/NIH)
Miller Lab (Rice University, Houston)
NIH Support: National Heart, Lung, and Blood Institute; National Institute of Biomedical Imaging and Bioengineering; National Institute of General Medical Sciences; Common Fund
Cool Videos: A Look Inside a Mouse’s Lung
Posted on by Dr. Francis Collins
If you have ever wondered what it is like to be an oxygen molecule inhaled through the lungs, here is your chance to find out! In this movie, we take a fantastic voyage through the slippery airways of the adult mouse lung.
We begin at the top in the main pipeline, called the bronchus, just below the trachea and wind through a system of increasingly narrow tubes. As you zoom through the airways, take note of the cilia (seen as goldish streaks); these tiny, hair-like structures move dust, germs, and mucus from smaller air passages to larger ones. Our quick trip concludes with a look into the alveoli — the air sacs where oxygen is delivered to red blood cells and carbon dioxide is removed and exhaled.


